When I first started my family history research over twenty years ago I began by compiling the family information in Word files. It soon became too complicated to keep track of all the different family lines in separate files and so I bought a family history program to record all the information. After researching all the available family history programs I settled on Family Historian, which has an excellent reputation and is well-suited for one-name study work. The program has stood me in good stead over the years. I use it to record my own family tree but also all the other Cruwys and Cruse trees I've researched as part of my one-name study.
When I first tested at AncestryDNA in 2012 I only had a very basic skeleton tree on Ancestry. I didn't get much out of the DNA test in the early years and it was another three years before Ancestry officially launched their test in the UK in 2015. Since then, the database has grown exponentially, allowing me to confirm much of my existing research and also to make some exciting new discoveries. I've found that there are distinct advantages to maintaining a detailed family tree on Ancestry. Having a publicly accessible tree means that I benefit from the Common Ancestor Hints and ThruLines. The more I build out the tree and build down the collateral lines the easier it becomes to work out the relationship to my matches.
I find that Ancestry's online tree-building platform is by far the easiest to use of all the available options. The powerful hint system makes it very easy to find records. Other family historians will often already have done research into a particular line and found all the relevant census and parish register entries which saves the hassle of spending a long time searching indexes to find the people I'm looking for, especially when the names have been horribly mistranscribed. Many people have generously shared photos as well as images of birth, marriage and death certificates which has been extremely helpful.
For all these reasons, over the last few years I've been slowly going through the process of trying to expand my tree on Ancestry by adding in all the research that I've done over the last twenty years which has been carefully stored in Family Historian. Rather than uploading a GEDCOM file and losing all the linked records and DNA matches I took the decision to curate my tree manually. I am gradually working through all the different branches of my tree adding census entries, uploading certificates and photos and linking them all to my tree. I've benefited greatly from the research that others have shared with me so I am effectively paying forwards the favour. This process has been invaluable in its own right. When I started out I was mostly working with indexed records and transcriptions. Increasingly records have been digitised and I am now able to link to census pages and original parish register records. I'm also making some very interesting new discoveries because of the consolidation process and because the hints often lead me to records which I hadn't previously discovered.
By adding sources to my tree I'm also hoping that my tree will have better exposure on Ancestry so that people can find my research. I'm also hoping that it will help to counter some of the mistakes I've found in some of the other online trees. Ancestry do not tell us how their algorithms work but it seems that the more sources you have linked to a person the more likely it is that your tree will appear in the hints list for other researchers.
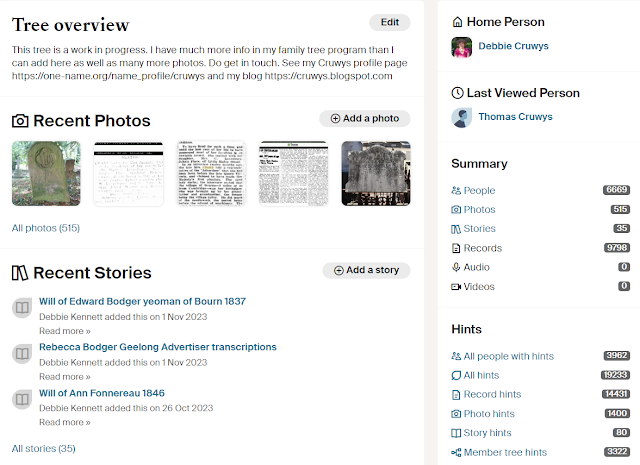
I've made good progress but there is still a lot more work to do. However, I'm pleased to report that I've completed the work on my own Cruwys family. My tree on Ancestry now encompasses my own Cruwys family from Winkleigh, the Cruwys Morchard tree, and the Mariansleigh tree. The Winkleigh and Cruwys Morchard trees link together in the mid-1400s. I know from the Y-DNA testing for my Cruwys DNA Project that the Mariansleigh Cruwyses are related to the Winkleigh and Cruwys Morchard families but I have been unable to document the connection. It does not help that the parish registers for the period in question have not survived. However, I have a second connection to the Mariansleigh Cruwyses through the Eastmond family so I was able to link them into one big tree. The Mariansleigh Cruwyses are by far the most numerous branch of the family so it took me some time to add all these records. If you're connected to any of these Cruwys families do check out my tree. You can find it on Ancestry here. If you have any additional information on these families I would be delighted to hear from you.
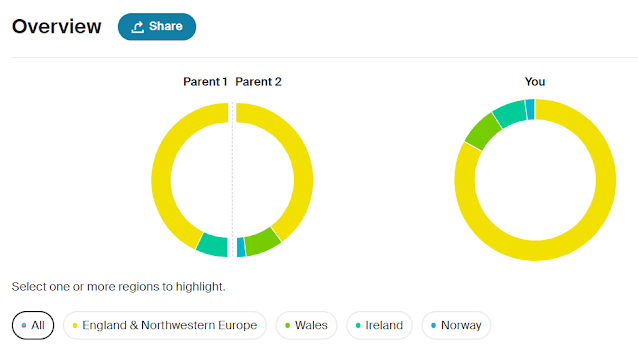
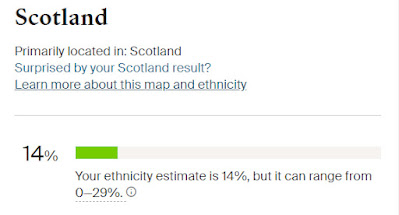
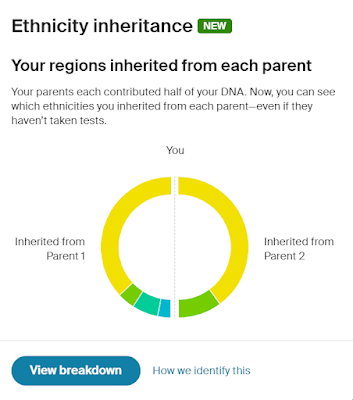
One of my next steps will be to build out trees on Ancestry for all the other Cruwys families I've researched as part of my one-name study. The Y-DNA project has shown that we have two distinct Cruwys groups which do not share a common ancestor and which belong to two completely different haplogroups. To make the work easier I am intending to upload a GEDCOM file for these other Cruwys families to a separate tree on Ancestry. I will then repeat the process of linking in all the census entries, parish registers and other records. I know it will take me some time to do this but it will be very worthwhile and will help to make my research more accessible.